Extending Similarity Network-Based Classifiers to the Non-Coding Genome and Deep Learning
Abstract
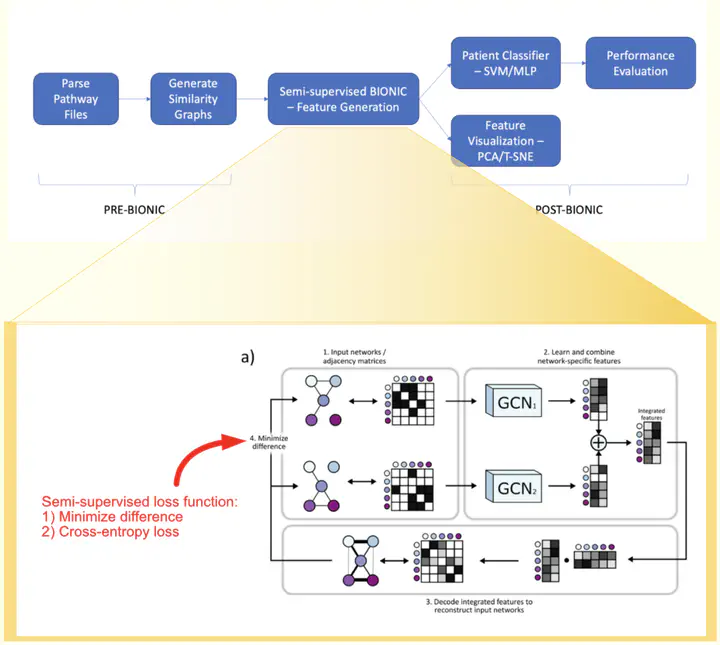
Similarity networks provide a useful framework for multi-modal data integration, suitable for applications such as gene function prediction and patient classification. We previously developed a supervised learning algorithm which converted heterogeneous patient data into the common space of patient similarity networks (PSN) and used these networks as input features2 (netDx.org). In addition to excellent classification performance and handling missing data, netDx provides interpretability by allowing users to group genes into pathway-level features. However, the pathway-based grouping approach is of limited value for genomic data outside coding regions. Moreover, the current framework has limited scalability in the number of nodes and networks and does not take advantage of improved discriminability available in the deep learning framework. Here, we describe two recent areas of work addressing these limitations. In the first, we classify binary survival in PFA ependymomas using tumour DNA methylomes organized with prior knowledge of brain tissue- and cell-specific expression, transcription factor binding sites and chromatin state. In the second, we extend a recently developed framework called BIONIC for multiple network integration based on graph convolutional networks, to classification. Developing an approach to score features for interpretability remains an active area of research.